Analysis of pupil data¶
There are many valid approaches to the analysis of pupillometry data, but the optimal approach will depend on the type of experiment being run and the research question in mind. Kelbsch et al. (2019) provide an informative view on standards in pupillometry, and there are some great examples of community-developed packages for streamlining pupillometry data analysis. Whilst it is always worth exploring the wider options that are available, PyPlr includes a set of pandas-reliant (you won’t regret learning pandas!) scripting tools for implementing a standard data processing pipeline that is optimised to work with some of the idiosynchrasies of Pupil Labs data. So far we have found these tools to be adequate for analysing our own data, but we welcome suggestions for improvements.
See here for a general introduction to analysing Pupil Labs pupillometry data. What follows is a typical PyPlr analysis pipeline with tools from pyplr.utils and pyplr.preproc.
Export with Pupil Player¶
The first step in our pipeline is to export the data using Pupil Player, making sure the required plugins (e.g., Annotation Player plugin) are enabled. Currently this must be done individually for each recording. Below is printed the file structure of a typical recording after export (more info on recording format here).
[1]:
from pyplr import utils
# Pupil Labs recording directory
rec_dir = '/Users/jtm/OneDrive - Nexus365/protocols/pipr_protocol/JTM'
utils.print_file_structure(rec_dir)
JTM/
.DS_Store
annotation.pldata
annotation_player.pldata
annotation_player_timestamps.npy
annotation_timestamps.npy
blinks.pldata
blinks_timestamps.npy
eye1.intrinsics
eye1.mp4
eye1_lookup.npy
eye1_timestamps.npy
fixations.pldata
fixations_timestamps.npy
gaze.pldata
gaze_timestamps.npy
info.player.json
notify.pldata
notify_timestamps.npy
pupil.pldata
pupil_timestamps.npy
surface_definitions_v01
user_info.csv
world.intrinsics
world.mp4
world_lookup.npy
world_timestamps.npy
analysis/
plr_extraction.png
plr_metrics.csv
plr_overall_metrics.png
processed.csv
pupil_processing.png
exports/
000/
annotations.csv
export_info.csv
gaze_positions.csv
pupil_gaze_positions_info.txt
pupil_positions.csv
world.mp4
world_timestamps.csv
world_timestamps.npy
pyplr_analysis/
offline_data/
tokens/
gaze_positions_consumer_Vis_Polyline.token
gaze_positions_producer_GazeFromRecording.token
pupil_positions_consumer_Pupil_From_Recording.token
pupil_positions_consumer_System_Timelines.token
pupil_positions_producer_Pupil_From_Recording.token
Load exported data¶
Now we can set up some constants and use pyplr.utils to get things moving. new_subject(...) returns a dictionary with the root directory, recording id, data directory and a newly made output directory for the analysis results. Then, passing the data directory to load_pupil(...), we load the specified columns from the pupil_positions.csv exported data.
[2]:
# Columns to load
use_cols = ['confidence',
'method',
'pupil_timestamp',
'eye_id',
'diameter_3d',
'diameter']
# Get a handle on a subject
s = utils.new_subject(
rec_dir, export='000', out_dir_nm='pyplr_analysis')
# Load pupil data
samples = utils.load_pupil(
s['data_dir'], eye_id='best', cols=use_cols)
samples
************************************************************
*************************** JTM ****************************
************************************************************
Loaded 48552 samples
[2]:
| eye_id | confidence | diameter | method | diameter_3d | |
|---|---|---|---|---|---|
| pupil_timestamp | |||||
| 85838.895658 | 1 | 0.977211 | 50.102968 | 3d c++ | 6.105771 |
| 85838.903656 | 1 | 0.997867 | 50.037086 | 3d c++ | 6.099682 |
| 85838.911644 | 1 | 0.998295 | 49.702628 | 3d c++ | 6.060059 |
| 85838.919551 | 1 | 0.997833 | 49.968637 | 3d c++ | 6.093759 |
| 85838.927504 | 1 | 0.998183 | 49.883191 | 3d c++ | 6.084404 |
| ... | ... | ... | ... | ... | ... |
| 86246.730916 | 1 | 0.818574 | 56.684975 | 3d c++ | 6.966634 |
| 86246.739307 | 1 | 0.947873 | 56.824819 | 3d c++ | 6.983579 |
| 86246.746889 | 1 | 0.969337 | 57.176728 | 3d c++ | 7.027931 |
| 86246.758588 | 1 | 0.956524 | 56.838490 | 3d c++ | 6.988390 |
| 86246.762987 | 1 | 0.927155 | 56.933094 | 3d c++ | 7.000156 |
48552 rows × 5 columns
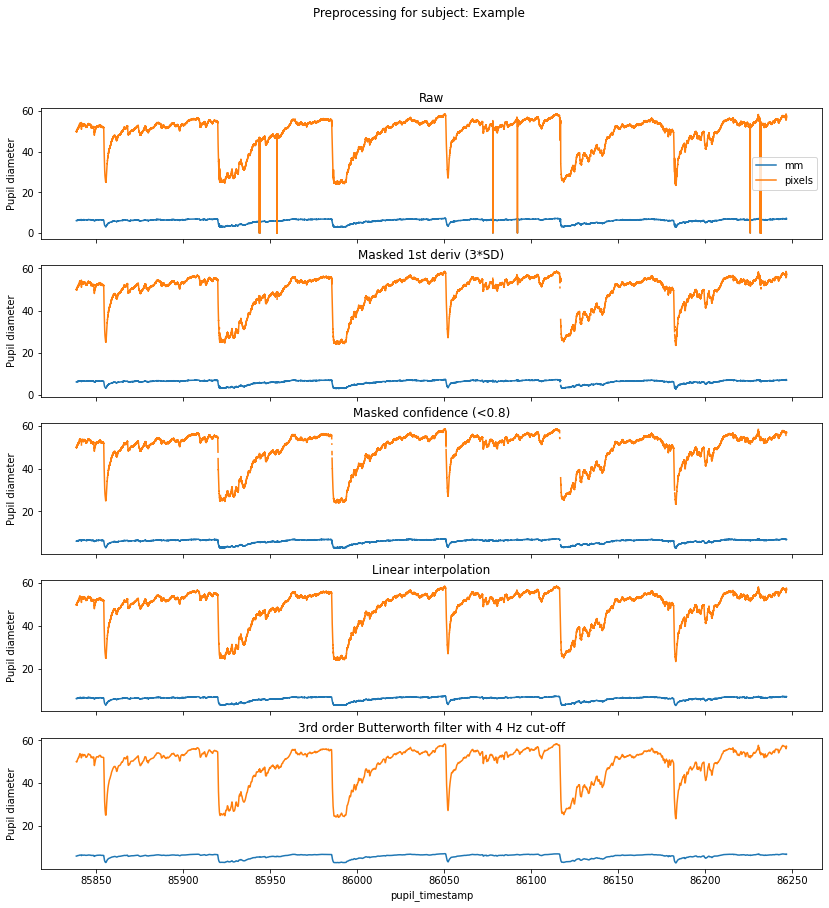
Preprocessing¶
Preprocessing pupillometry data is relatively straight-forward and typically involves dealing with signal loss due to eye blinks and smoothing out any high frequency noise. There are sophisticated algorithms for detecting blinks in a pupil time course, but Pupil Player has its own Blink Detection plugin that detects blinks based on rapid changes in the confidence metric. Whilst you can export these blink events and use the timestamps to index and mask the pupil timecourse, we find that it is effective simply to mask the pupil data with thresholds on the first derivative and confidence metric, and then to follow up with linear interpolation and smoothing.
[3]:
from pyplr import graphing
from pyplr import preproc
# Sampling frequency
SAMPLE_RATE = 120
# Pupil columns to analyse
pupil_cols = ['diameter_3d', 'diameter']
# Make figure for processing
f, axs = graphing.pupil_preprocessing(nrows=5, subject='Example')
# Plot the raw data
samples[pupil_cols].plot(title='Raw', ax=axs[0], legend=True)
axs[0].legend(loc='center right', labels=['mm', 'pixels'])
# Mask first derivative
samples = preproc.mask_pupil_first_derivative(
samples, threshold=3.0, mask_cols=pupil_cols)
samples[pupil_cols].plot(
title='Masked 1st deriv (3*SD)', ax=axs[1], legend=False)
# Mask confidence
samples = preproc.mask_pupil_confidence(
samples, threshold=0.8, mask_cols=pupil_cols)
samples[pupil_cols].plot(
title='Masked confidence (<0.8)', ax=axs[2], legend=False)
# Interpolate
samples = preproc.interpolate_pupil(
samples, interp_cols=pupil_cols)
samples[pupil_cols].plot(
title='Linear interpolation', ax=axs[3], legend=False)
# Smooth
samples = preproc.butterworth_series(
samples, fields=pupil_cols, filt_order=3,
cutoff_freq=4/(SAMPLE_RATE/2))
samples[pupil_cols].plot(
title='3rd order Butterworth filter with 4 Hz cut-off',
ax=axs[4], legend=False);
Extraction¶
Having cleaned the timecourse we need to extract the events of interest. This requires the annotation events sent during the recording, which have the timestamps needed for extraction. These can be loaded with utlis.load_annotations(...).
[4]:
events = utils.load_annotations(s['data_dir'])
events
Loaded 6 events
[4]:
| index | label | duration | color | creation_time | creator | protocol | pulse_duration | pulse_spec | |
|---|---|---|---|---|---|---|---|---|---|
| timestamp | |||||||||
| 85854.348667 | 1835 | LIGHT_ON | NaN | red | 2020-11-10 09:39:10.888989 | jtm | pulse | 1000 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 1979] |
| 85919.820362 | 9629 | LIGHT_ON | NaN | blue | 2020-11-10 09:39:10.882007 | jtm | pulse | 1000 | [0, 0, 0, 2500, 0, 0, 0, 0, 0, 0] |
| 85985.280630 | 17422 | LIGHT_ON | NaN | blue | 2020-11-10 09:39:10.882007 | jtm | pulse | 1000 | [0, 0, 0, 2500, 0, 0, 0, 0, 0, 0] |
| 86050.768628 | 25217 | LIGHT_ON | NaN | red | 2020-11-10 09:39:10.888989 | jtm | pulse | 1000 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 1979] |
| 86116.228025 | 33008 | LIGHT_ON | NaN | blue | 2020-11-10 09:39:10.882007 | jtm | pulse | 1000 | [0, 0, 0, 2500, 0, 0, 0, 0, 0, 0] |
| 86181.688172 | 40801 | LIGHT_ON | NaN | red | 2020-11-10 09:39:10.888989 | jtm | pulse | 1000 | [0, 0, 0, 0, 0, 0, 0, 0, 0, 1979] |
Now we can pass our samples and events to utils.extract(...) along with some parameters defining the number of samples to extract and how much to offset the data relative to the event. Here we extract-65 second epochs and offset by a 5 second baseline period.
[5]:
# Number of samples to extract and which sample
# should mark the onset of the event
DURATION = 7800
ONSET_IDX = 600
# Extract the event ranges
ranges = utils.extract(
samples,
events,
offset=-ONSET_IDX,
duration=DURATION,
borrow_attributes=['color'])
ranges
Extracted ranges for 6 events
[5]:
| eye_id | confidence | diameter | method | diameter_3d | interpolated | orig_idx | color | ||
|---|---|---|---|---|---|---|---|---|---|
| event | onset | ||||||||
| 0 | 0 | 1.0 | 0.998989 | 48.404937 | 3d c++ | 5.990967 | 0 | 85849.311553 | red |
| 1 | 1.0 | 0.998972 | 48.436117 | 3d c++ | 5.994949 | 0 | 85849.319757 | red | |
| 2 | 1.0 | 0.999134 | 48.469848 | 3d c++ | 5.999237 | 0 | 85849.327708 | red | |
| 3 | 1.0 | 0.998426 | 48.505896 | 3d c++ | 6.003802 | 0 | 85849.335675 | red | |
| 4 | 1.0 | 0.998895 | 48.544031 | 3d c++ | 6.008612 | 0 | 85849.343669 | red | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5 | 7795 | 1.0 | 0.922093 | 54.375155 | 3d c++ | 6.707360 | 0 | 86242.127040 | red |
| 7796 | 1.0 | 0.977674 | 54.338056 | 3d c++ | 6.703963 | 0 | 86242.135026 | red | |
| 7797 | 1.0 | 0.933031 | 54.304633 | 3d c++ | 6.701016 | 0 | 86242.142965 | red | |
| 7798 | 1.0 | 0.970404 | 54.274990 | 3d c++ | 6.698505 | 0 | 86242.150957 | red | |
| 7799 | 1.0 | 0.903852 | 54.249187 | 3d c++ | 6.696413 | 0 | 86242.162996 | red |
46800 rows × 8 columns
utils.extract(...) returns a pandas DataFrame with a two-level (event, onset) hierarchichal index. The original index to the data is preserved in a column along with any attributes borrowed from the annotations. Now is a good time to create additional columns expressing the pupil data as percent signal change from baseline.
[6]:
# Calculate baselines
baselines = ranges.loc[:, range(0, ONSET_IDX), :].mean(level=0)
# New columns for percent signal change
ranges = preproc.percent_signal_change(
ranges, baselines, pupil_cols)
ranges
[6]:
| eye_id | confidence | diameter | method | diameter_3d | interpolated | orig_idx | color | diameter_3d_pc | diameter_pc | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| event | onset | ||||||||||
| 0 | 0 | 1.0 | 0.998989 | 48.404937 | 3d c++ | 5.990967 | 0 | 85849.311553 | red | -6.907039 | -7.009474 |
| 1 | 1.0 | 0.998972 | 48.436117 | 3d c++ | 5.994949 | 0 | 85849.319757 | red | -6.845173 | -6.949574 | |
| 2 | 1.0 | 0.999134 | 48.469848 | 3d c++ | 5.999237 | 0 | 85849.327708 | red | -6.778534 | -6.884774 | |
| 3 | 1.0 | 0.998426 | 48.505896 | 3d c++ | 6.003802 | 0 | 85849.335675 | red | -6.707607 | -6.815521 | |
| 4 | 1.0 | 0.998895 | 48.544031 | 3d c++ | 6.008612 | 0 | 85849.343669 | red | -6.632865 | -6.742260 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5 | 7795 | 1.0 | 0.922093 | 54.375155 | 3d c++ | 6.707360 | 0 | 86242.127040 | red | 8.891820 | 8.796684 |
| 7796 | 1.0 | 0.977674 | 54.338056 | 3d c++ | 6.703963 | 0 | 86242.135026 | red | 8.836673 | 8.722455 | |
| 7797 | 1.0 | 0.933031 | 54.304633 | 3d c++ | 6.701016 | 0 | 86242.142965 | red | 8.788822 | 8.655579 | |
| 7798 | 1.0 | 0.970404 | 54.274990 | 3d c++ | 6.698505 | 0 | 86242.150957 | red | 8.748060 | 8.596268 | |
| 7799 | 1.0 | 0.903852 | 54.249187 | 3d c++ | 6.696413 | 0 | 86242.162996 | red | 8.714104 | 8.544640 |
46800 rows × 10 columns
Plotting and parametrisation¶
It is common practice when analysing the PLR to describe it in terms of paremeters relating to time, velocity and acceleration. So once you have an array of data representing a pupil’s response to light, be it from a single trial or an average of multiple trials, simply pass it to pyplr.plr.PLR along with some basic parameters.
[7]:
from pyplr.plr import PLR
average_plr = ranges.mean(level=1)['diameter_3d'].to_numpy()
plr = PLR(average_plr,
sample_rate=SAMPLE_RATE,
onset_idx=ONSET_IDX,
stim_duration=1)
Now you can do a quick plot with options to display the velocity and acceleration profiles and the derived parameters:
[8]:
fig = plr.plot(vel=True, acc=True, print_params=True)
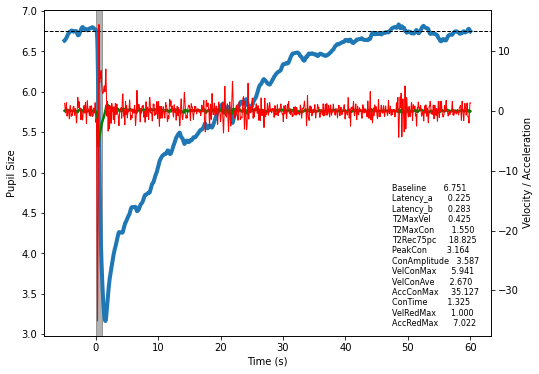
Or simply access the parameters as a DataFrame.
[9]:
params = plr.parameters()
params
[9]:
| value | |
|---|---|
| Baseline | 6.751031 |
| Latency_a | 0.225000 |
| Latency_b | 0.283333 |
| T2MaxVel | 0.425000 |
| T2MaxCon | 1.550000 |
| T2Rec75pc | 18.825000 |
| PeakCon | 3.164023 |
| ConAmplitude | 3.587008 |
| VelConMax | 5.940593 |
| VelConAve | 2.670336 |
| AccConMax | 35.127363 |
| ConTime | 1.325000 |
| VelRedMax | 0.999670 |
| AccRedMax | 7.021978 |